Novel Fetal Hemoglobin Modifying Loci Revealed with Genome Wide Studies of Sickle Cell Disease Patients from Cameroon, and Global Metanalysis
Content
- Load required r package
- Clinical datasets
- Summary of Cameroon data
- Summary of Tanzania data
- HbF normalization
- HbF distribution before and after normalization
Load require r package
library(tidyverse)
Clinical datasets
cm <- read_table("../clinicaldata/cm_clinical_data.tsv")
tz <- read_table("../clinicaldata/tz_clinical_data.tsv")
Summary of Cameroon data
Variables
cm %>% names()
## [1] "FID" "IID" "AGE"
## [4] "SEX" "HbF" "HbS"
## [7] "Hb_Phenotype" "Hb" "Phenotypes"
## [10] "Genotypes" "Haplotypes" "athal"
## [13] "Curated_Age_yrs" "VOC.year" "Consultation.year"
## [16] "Hosp" "Stroke" "HbA2_percentage"
## [19] "RBC" "MCV.fl." "MCHC"
## [22] "WBC" "Lymp" "Mono"
## [25] "Platelets" "Curated_penotype" "Sample_name_comment"
Summary statistics
Quantitative/Continuous variables
variables <- c(
"AGE", # Age in years
"HbS", # Proportion of HbS
"HbF", # Proportion of fetal haemoglobin
"HbA2_percentage", # Proportion of HbA2
"Consultation.year", # Number of consultations per year
"MCHC", # Mean corpuscular haemoglobin concentration
"Mono", # Monocyte count (x10^9/l)
"Hosp", # Number of hospitalizations per year
"RBC", # Red blood cell count (x10^12/l)
"WBC", # White blood cell count (x10^9/l)
"Platelets", # Platelet count (x10^9/l)
"VOC.year", # Number of vaso occlusive crises per year
"MCV.fl.", # Mean cell volume
"Lymp" # Lymphocyte count (x10^9/l)
)
cm %>%
select(variables) %>%
summary()
## AGE HbS HbF HbA2_percentage
## Min. : 5.00 Min. :19.60 Min. : 0.700 Min. : 0.000
## 1st Qu.:10.00 1st Qu.:80.30 1st Qu.: 4.700 1st Qu.: 2.700
## Median :16.00 Median :86.00 Median : 8.500 Median : 3.200
## Mean :18.05 Mean :83.56 Mean : 9.892 Mean : 3.324
## 3rd Qu.:24.00 3rd Qu.:90.70 3rd Qu.:13.600 3rd Qu.: 3.800
## Max. :66.00 Max. :96.50 Max. :37.400 Max. :18.200
## NA's :3
## Consultation.year MCHC Mono Hosp
## Min. : 0.000 Min. : 3.80 Min. :0.000 Min. : 0.000
## 1st Qu.: 0.000 1st Qu.:25.90 1st Qu.:0.700 1st Qu.: 1.000
## Median : 1.000 Median :31.00 Median :1.000 Median : 1.000
## Mean : 2.049 Mean :29.21 Mean :1.161 Mean : 1.974
## 3rd Qu.: 2.000 3rd Qu.:33.00 3rd Qu.:1.400 3rd Qu.: 2.000
## Max. :15.000 Max. :54.20 Max. :9.400 Max. :24.000
## NA's :805 NA's :17 NA's :14 NA's :59
## RBC WBC Platelets VOC.year
## Min. :0.84 Min. : 0.400 Min. : 22.0 Min. : 0.000
## 1st Qu.:2.29 1st Qu.: 8.607 1st Qu.: 310.0 1st Qu.: 1.000
## Median :2.70 Median :11.200 Median : 401.0 Median : 2.000
## Mean :2.75 Mean :11.819 Mean : 415.4 Mean : 3.189
## 3rd Qu.:3.12 3rd Qu.:14.200 3rd Qu.: 498.0 3rd Qu.: 4.000
## Max. :7.70 Max. :49.800 Max. :1185.0 Max. :48.000
## NA's :15 NA's :14 NA's :13 NA's :29
## MCV.fl. Lymp
## Min. : 59.0 Min. : 0.100
## 1st Qu.: 81.0 1st Qu.: 3.570
## Median : 87.0 Median : 4.700
## Mean : 87.1 Mean : 5.186
## 3rd Qu.: 94.0 3rd Qu.: 6.210
## Max. :132.0 Max. :22.600
## NA's :14 NA's :13
Categorigal variables
cm %>%
select(SEX) %>%
table() %>%
as.data.frame() %>%
na.omit() %>%
mutate(Prop = (Freq/sum(Freq))*100) -> sex # Sex
sex
## . Freq Prop
## 1 F 657 52.56
## 2 M 593 47.44
colnames(sex) <- c(
"cat",
"freq",
"prop"
)
par(
mar = c(4,4,3,4)
)
barplot(
sex$prop,
ylim = c(0, 60),
col = c(2,4)
)
text(
x = c(0.7,1.9),
y = 30,
labels = c("Female", "Male")
)
mtext(
text = "Proportion (%)",
side = 2,
line = 3
)
mtext(
text = "Sex",
side = 1,
line = 1
)
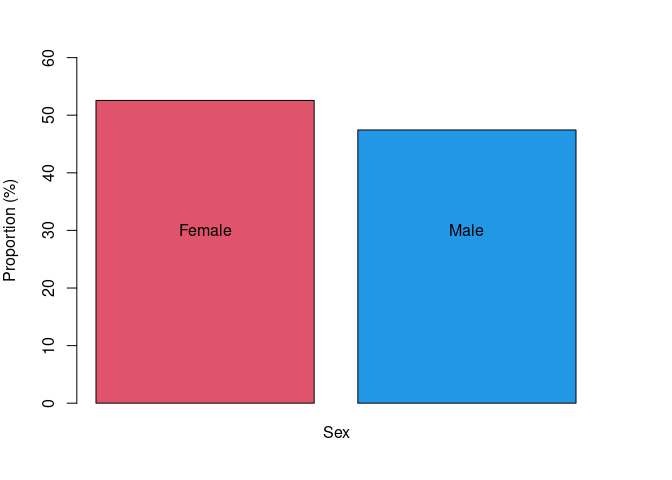
Figue 1. Proportion of females and males in the discovery dataset of Cameroonian inidividuals
# Number of patients with at least one overt stroke
cm %>%
select(Stroke) %>%
table()
## .
## No Yes
## 1195 46
# 3.7 kb HbA1/HbA2 alpha thalassaemia deletion
cm %>%
select(athal) %>%
table()
## .
## α3.7/α3.7 αα/α3.7 αα/αα
## 18 101 286
# HBB gene cluster haplotypes
cm %>%
select(Haplotypes) %>%
table()
## .
## Atypical BEN BEN/AI BEN/Atypical BEN/CAM BEN/CAR
## 3 331 2 26 99 2
## CAM CAM/AI CAM/Atypical CAM/SEN CAR CAR/Atypical
## 174 1 8 1 1 1
## CAR/CAM SEN
## 1 4
CAM = CAM/CAM, BEN = BEN/BEN, SEN = SEN/SEN, CAR = CAR/CAR, Atypical = Atypical/Atypical
Summary of Tanzania data
Variables
tz %>% names()
## [1] "FID" "IID" "AGE" "HbF" "SEX"
Summary statistics
Quantitative/Continuous variables
variables <- c(
"AGE",
"HbF"
)
tz %>%
select(variables) %>%
summary()
## AGE HbF
## Min. : 5.021 Min. : 0.200
## 1st Qu.: 7.876 1st Qu.: 2.600
## Median :11.069 Median : 4.800
## Mean :13.288 Mean : 5.748
## 3rd Qu.:16.298 3rd Qu.: 7.800
## Max. :44.310 Max. :32.500
Categorical variable
tz %>%
mutate(sex.updated = ifelse(SEX == 1, "M", ifelse(SEX == 2, "F", NA))) %>%
select(sex.updated) %>%
table() %>%
as.data.frame() %>%
na.omit() %>%
mutate(Prop = (Freq/sum(Freq))*100) -> sex # Sex
sex
## . Freq Prop
## 1 F 539 52.895
## 2 M 480 47.105
colnames(sex) <- c(
"cat",
"freq",
"prop"
)
par(
mar = c(4,4,3,4)
)
barplot(
sex$prop,
ylim = c(0, 60),
col = c(2,4)
)
text(
x = c(0.7,1.9),
y = 30,
labels = c("Female", "Male")
)
mtext(
text = "Proportion (%)",
side = 2,
line = 3
)
mtext(
text = "Sex",
side = 1,
line = 1
)
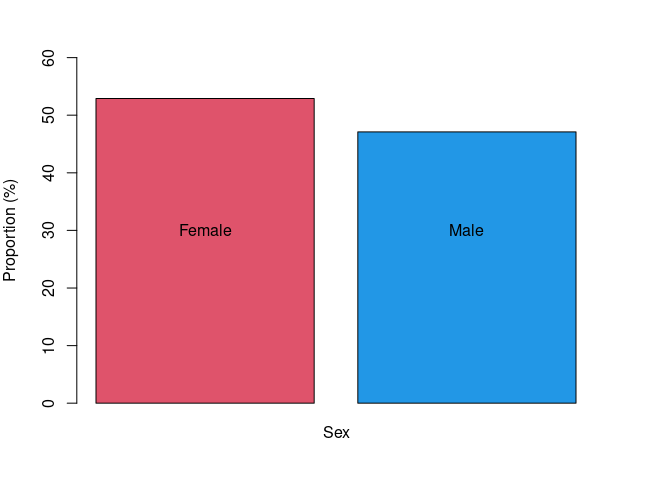
Figure 2. Proportion of females and males in the replication dataset of Tanzanian inidividuals
HbF normalization (Cubic root)
Cameroon
cm %>%
mutate(normalizedHbF = HbF^(1/3)) -> cm.hbf.norm
write.table(
cm.hbf.norm,
"../clinicaldata/cm_clinical_data_updated.tsv",
col.names = T,
row.names = F,
quote = F,
sep = "\t"
)
cm.hbf.norm %>%
select(HbF, normalizedHbF) %>%
head()
## # A tibble: 6 × 2
## HbF normalizedHbF
## <dbl> <dbl>
## 1 31.6 3.16
## 2 6.4 1.86
## 3 12.6 2.33
## 4 8.8 2.06
## 5 17.5 2.60
## 6 17.8 2.61
Tanzania
# Tanzania
tz %>%
mutate(normalizedHbF = HbF^(1/3)) -> tz.hbf.norm
write.table(
tz.hbf.norm,
"../clinicaldata/tz_clinical_data_updated.tsv",
col.names = T,
row.names = F,
quote = F,
sep = "\t"
)
tz.hbf.norm %>%
select(HbF, normalizedHbF) %>%
head()
## # A tibble: 6 × 2
## HbF normalizedHbF
## <dbl> <dbl>
## 1 0.2 0.585
## 2 0.3 0.669
## 3 0.3 0.669
## 4 0.3 0.669
## 5 0.3 0.669
## 6 0.4 0.737
HbF distribution before and after normalization
cm.hbf.den <- density(
cm.hbf.norm$HbF
)
cm.nhbf.den <- density(
cm.hbf.norm$normalizedHbF
)
tz.hbf.den <- density(
tz.hbf.norm$HbF
)
tz.nhbf.den <- density(
tz.hbf.norm$normalizedHbF
)
par(
mar = c(4,4,3,4)
)
plot(
cm.hbf.den,
main = "Cameroon HbF"
)
polygon(
cm.hbf.den,
col = 2
)
plot(
cm.nhbf.den,
ylab = "",
main = "Cameroon normalized HbF"
)
polygon(
cm.nhbf.den,
col=5
)
plot(
tz.hbf.den,
main = "Tanzania HbF"
)
polygon(
tz.hbf.den,
col = 2
)
plot(
tz.nhbf.den,
ylab = "",
main = "Tanzania normalized HbF"
)
polygon(
tz.nhbf.den,
col = 5
)

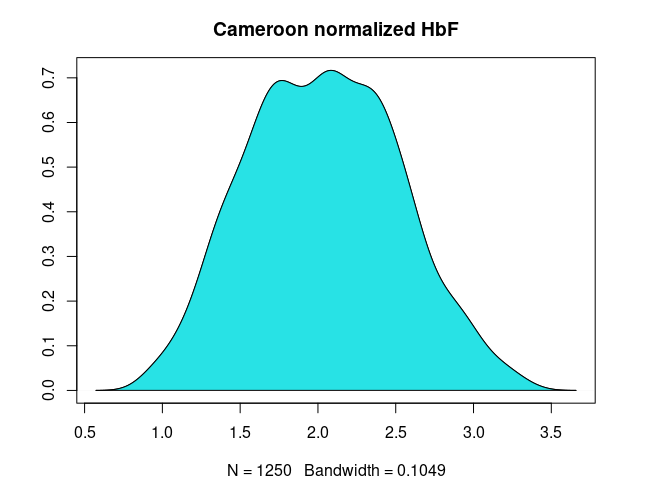
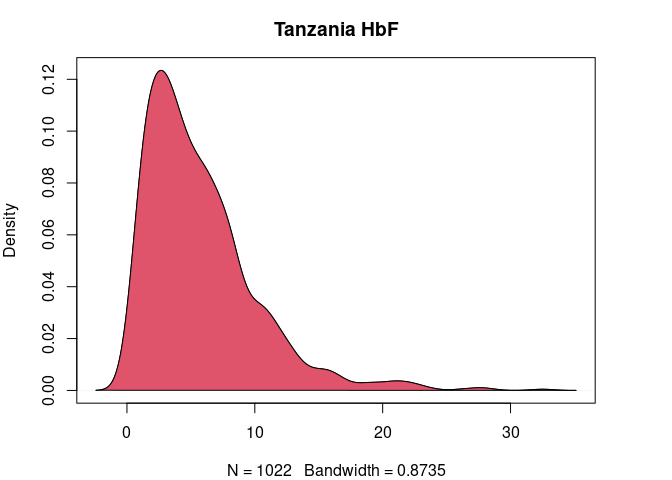
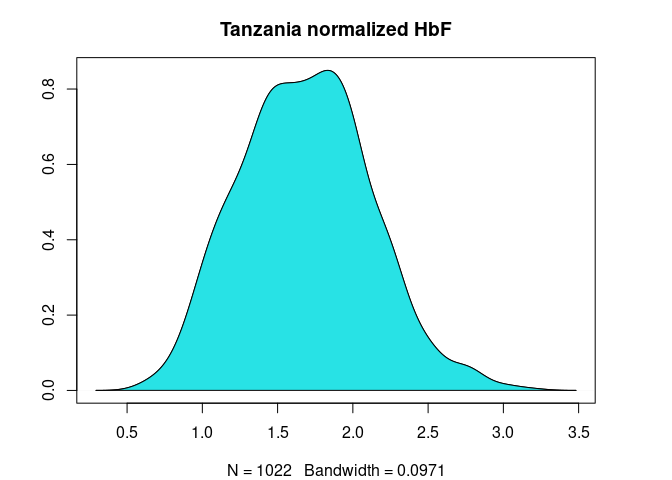
Figure 3. HbF distribution in Cameroonian and Tanzanian study participants before and after normalization